Characterization of the Bacterial Composition of 47 Fermented Foods in Sweden
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Fermented Foods
2.2. Preparing Fermented Foods in Home Environments
2.3. Sample Preparation and Handling
2.4. Microbiota Analysis
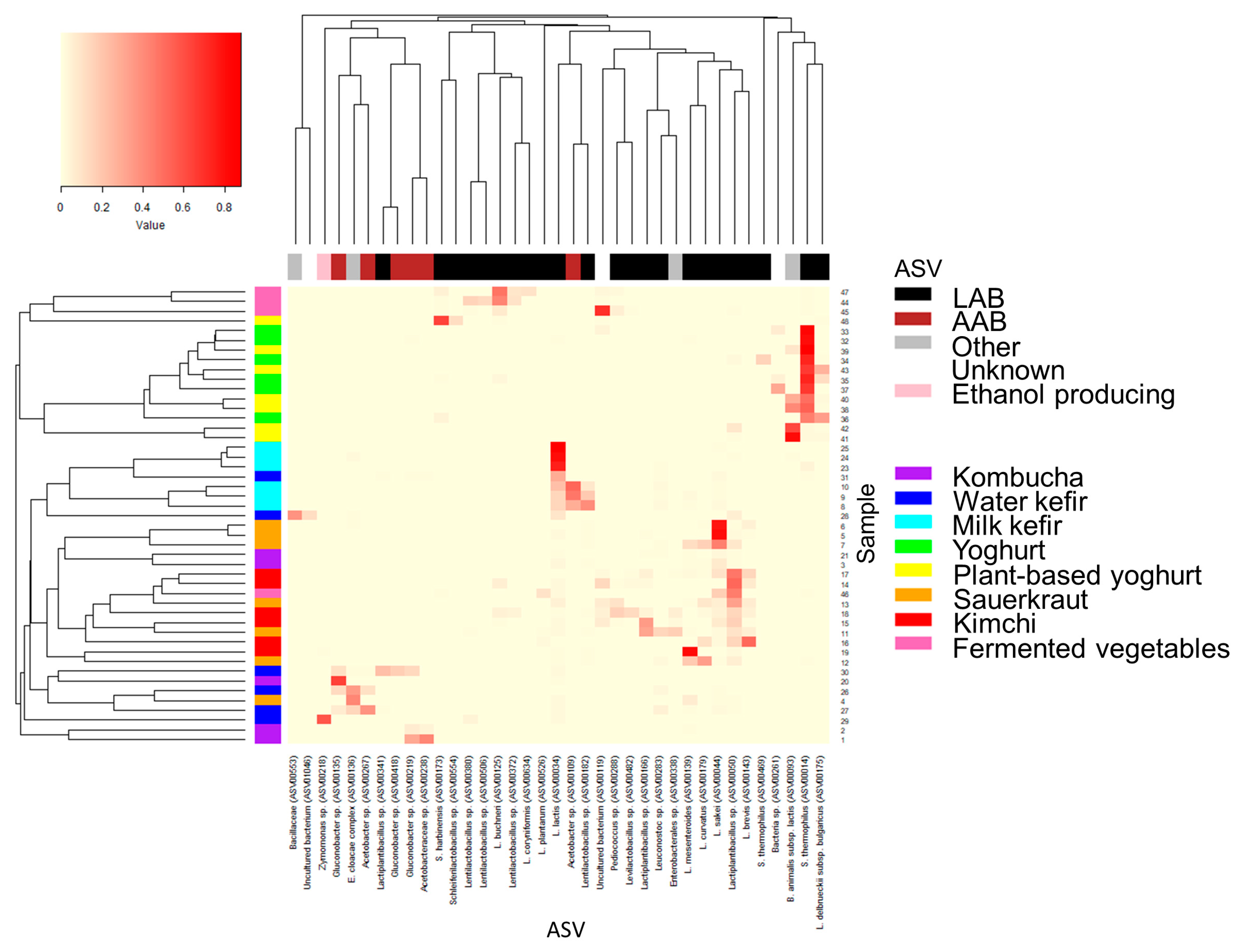
2.5. Taxonomic Assignment for Individual ASV with High Relative Abundance
2.6. Statistical Analysis
3. Results and Discussion
3.1. Alpha-Diversity and Richness Differed by Fermented Food Group
3.2. Dissimilarities in the Bacterial Community across Fermented Foods and Food Groups
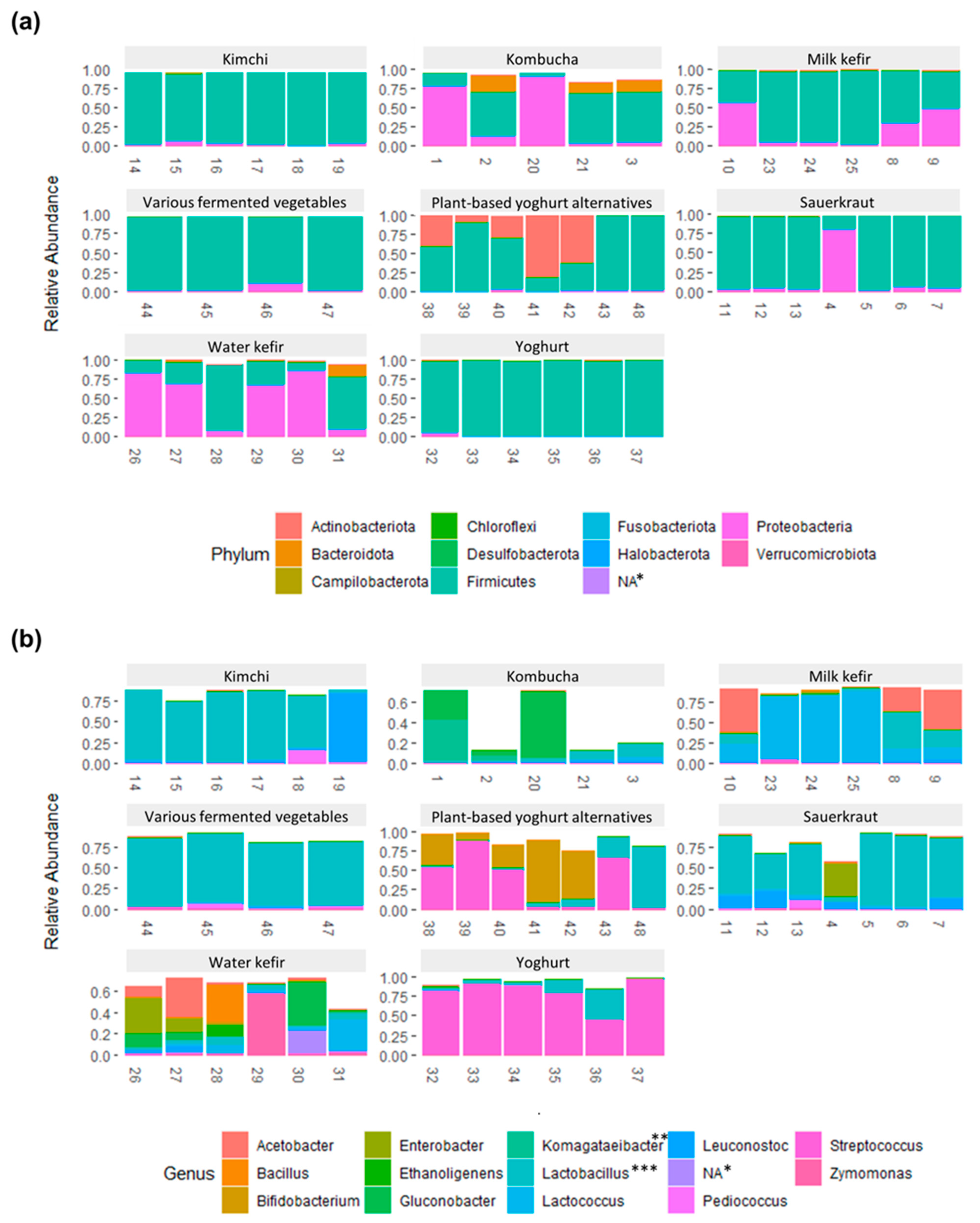
3.3. The Bacterial Composition of the Fermented Foods and Food Groups
3.4. Bacterial Composition and Variability within Each Fermented Food Group
3.4.1. Kombucha
3.4.2. Water Kefir
3.4.3. Milk Kefir
3.4.4. Yoghurt
3.4.5. Plant-Based Alternatives to Yoghurt
3.4.6. Sauerkraut
3.4.7. Kimchi
3.4.8. Various Fermented Vegetables
3.5. Final Remarks
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marco, M.L.; Sanders, M.E.; Ganzle, M.; Arrieta, M.C.; Cotter, P.D.; De Vuyst, L.; Hill, C.; Holzapfel, W.; Lebeer, S.; Merenstein, D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 196–208. [Google Scholar] [CrossRef]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of Microorganisms in Global Fermented Foods and Beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented foods in a global age: East meets West. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef] [PubMed]
- Voidarou, C.; Antoniadou, M.; Rozos, G.; Tzora, A.; Skoufos, I.; Varzakas, T.; Lagiou, A.; Bezirtzoglou, E. Fermentative Foods: Microbiology, Biochemistry, Potential Human Health Benefits and Public Health Issues. Foods 2020, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Gille, D.; Schmid, A.; Walther, B.; Vergeres, G. Fermented Food and Non-Communicable Chronic Diseases: A Review. Nutrients 2018, 10, 448. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, M.; O’Sullivan, O.; Cotter, P.D.; Sinderen, D.V.; Kenny, J.G. The Application of Metagenomics to Study Microbial Communities and Develop Desirable Traits in Fermented Foods. Foods 2022, 11, 3297. [Google Scholar] [CrossRef]
- Park, E.J.; Chun, J.; Cha, C.J.; Park, W.S.; Jeon, C.O.; Bae, J.W. Bacterial community analysis during fermentation of ten representative kinds of kimchi with barcoded pyrosequencing. Food Microbiol. 2012, 30, 197–204. [Google Scholar] [CrossRef]
- Kergourlay, G.; Taminiau, B.; Daube, G.; Champomier Verges, M.C. Metagenomic insights into the dynamics of microbial communities in food. Int. J. Food Microbiol. 2015, 213, 31–39. [Google Scholar] [CrossRef]
- Zhang, Z.; Han, Z.; Wu, Y.; Jiang, S.; Ma, C.; Zhang, Y.; Zhang, J. Metagenomics assembled genome scale analysis revealed the microbial diversity and genetic polymorphism of Lactiplantibacillus plantarum in traditional fermented foods of Hainan, China. Food Res. Int. 2021, 150, 110785. [Google Scholar] [CrossRef]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequence-based analysis of the microbial composition of water kefir from multiple sources. FEMS Microbiol. Lett. 2013, 348, 79–85. [Google Scholar] [CrossRef]
- Rahmani, B.; Alimadadi, N.; Attaran, B.; Nasr, S. Yeasts from Iranian traditional milk kefir samples: Isolation, molecular identification and their potential probiotic properties. Lett. Appl. Microbiol. 2022, 75, 1264–1274. [Google Scholar] [CrossRef]
- Lee, D.Y.; Park, S.H.; Park, S.E.; Kim, E.J.; Kim, H.W.; Seo, S.H.; Cho, K.M.; Kwon, S.J.; Whon, T.W.; Min, S.G.; et al. Comprehensive elucidation of the terroir of Korean kimchi through the study of recipes, metabolites, microbiota, and sensory characteristics. Food Res. Int. 2023, 166, 112614. [Google Scholar] [CrossRef]
- Wuyts, S.; Van Beeck, W.; Oerlemans, E.F.M.; Wittouck, S.; Claes, I.J.J.; De Boeck, I.; Weckx, S.; Lievens, B.; De Vuyst, L.; Lebeer, S. Carrot Juice Fermentations as Man-Made Microbial Ecosystems Dominated by Lactic Acid Bacteria. Appl. Environ. Microbiol. 2018, 84, e00134-18. [Google Scholar] [CrossRef]
- Van Reckem, E.; Geeraerts, W.; Charmpi, C.; Van der Veken, D.; De Vuyst, L.; Leroy, F. Exploring the Link Between the Geographical Origin of European Fermented Foods and the Diversity of Their Bacterial Communities: The Case of Fermented Meats. Front. Microbiol. 2019, 10, 2302. [Google Scholar] [CrossRef] [PubMed]
- Ashaolu, T.J.; Reale, A. A Holistic Review on Euro-Asian Lactic Acid Bacteria Fermented Cereals and Vegetables. Microorganisms 2020, 8, 1176. [Google Scholar] [CrossRef]
- De Jong, M.; Alekseeva, A.Y.; Miraji, K.F.; Phiri, S.; Linnemann, A.R.; Schoustra, S.E. Environmental Selection Shapes Bacterial Community Composition in Traditionally Fermented Maize-Based Foods from Benin, Tanzania and Zambia. Microorganisms 2022, 10, 1354. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, A.; Cheng, L.; Chen, Q.; Li, J.; Xu, Y.; Huo, D. Deep Shotgun metagenomic and 16S rRNA analysis revealed the microbial diversity of lactic acid bacteria in traditional fermented foods of eastern Hainan, China. Food Funct. 2022, 13, 12938–12952. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O'Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Beule, L.; Karlovsky, P. Improved normalization of species count data in ecology by scaling with ranked subsampling (SRS): Application to microbial communities. PeerJ 2020, 8, e9593. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, M.P. Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, Y.; Hong, H. Classification of acetic acid bacteria and their acid resistant mechanism. AMB Express 2021, 11, 29. [Google Scholar] [CrossRef]
- Coelho, R.M.D.; Almeida, A.L.d.; Amaral, R.Q.G.d.; Mota, R.N.d.; Sousa, P.H.M.d. Kombucha: Review. Int. J. Gastron. Food Sci. 2020, 22, 100272. [Google Scholar] [CrossRef]
- Lynch, K.M.; Wilkinson, S.; Daenen, L.; Arendt, E.K. An update on water kefir: Microbiology, composition and production. Int. J. Food Microbiol. 2021, 345, 109128. [Google Scholar] [CrossRef]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93, fix048. [Google Scholar] [CrossRef]
- Kaashyap, M.; Cohen, M.; Mantri, N. Microbial Diversity and Characteristics of Kombucha as Revealed by Metagenomic and Physicochemical Analysis. Nutrients 2021, 13, 4446. [Google Scholar] [CrossRef]
- Huang, X.; Xin, Y.; Lu, T. A systematic, complexity-reduction approach to dissect the kombucha tea microbiome. Elife 2022, 11, e76401. [Google Scholar] [CrossRef]
- Wang, B.; Rutherfurd-Markwick, K.; Zhang, X.X.; Mutukumira, A.N. Isolation and characterisation of dominant acetic acid bacteria and yeast isolated from Kombucha samples at point of sale in New Zealand. Curr. Res. Food Sci. 2022, 5, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lagishetty, V.; Kurnia, P.; Henning, S.M.; Ahdoot, A.I.; Jacobs, J.P. Microbial and Chemical Profiles of Commercial Kombucha Products. Nutrients 2022, 14, 670. [Google Scholar] [CrossRef] [PubMed]
- Fiorda, F.A.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Rakshit, S.K.; Pagnoncelli, M.G.B.; Vandenberghe, L.P.S.; Soccol, C.R. Microbiological, biochemical, and functional aspects of sugary kefir fermentation—A review. Food Microbiol. 2017, 66, 86–95. [Google Scholar] [CrossRef]
- Laureys, D.; Leroy, F.; Vandamme, P.; De Vuyst, L. Backslopping Time, Rinsing of the Grains During Backslopping, and Incubation Temperature Influence the Water Kefir Fermentation Process. Front. Microbiol. 2022, 13, 871550. [Google Scholar] [CrossRef] [PubMed]
- Nejati, F.; Junne, S.; Neubauer, P. A Big World in Small Grain: A Review of Natural Milk Kefir Starters. Microorganisms 2020, 8, 192. [Google Scholar] [CrossRef]
- Alraddadi, F.A.J.; Ross, T.; Powell, S.M. Evaluation of the microbial communities in kefir grains and kefir over time. Int. Dairy J. 2023, 136, 105490. [Google Scholar] [CrossRef]
- Fisberg, M.; Machado, R. History of yogurt and current patterns of consumption. Nutr. Rev. 2015, 73, 4–7. [Google Scholar] [CrossRef]
- Montemurro, M.; Pontonio, E.; Coda, R.; Rizzello, C.G. Plant-Based Alternatives to Yogurt: State-of-the-Art and Perspectives of New Biotechnological Challenges. Foods 2021, 10, 316. [Google Scholar] [CrossRef]
- Mudoor Sooresh, M.; Willing, B.P.; Bourrie, B.C.T. Opportunities and Challenges of Understanding Community Assembly in Spontaneous Food Fermentation. Foods 2023, 12, 673. [Google Scholar] [CrossRef]
- Touret, T.; Oliveira, M.; Semedo-Lemsaddek, T. Putative probiotic lactic acid bacteria isolated from sauerkraut fermentations. PLoS ONE 2018, 13, e0203501. [Google Scholar] [CrossRef]
- Zabat, M.A.; Sano, W.H.; Wurster, J.I.; Cabral, D.J.; Belenky, P. Microbial Community Analysis of Sauerkraut Fermentation Reveals a Stable and Rapidly Established Community. Foods 2018, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Gaudioso, G.; Weil, T.; Marzorati, G.; Solovyev, P.; Bontempo, L.; Franciosi, E.; Bertoldi, L.; Pedrolli, C.; Tuohy, K.M.; Fava, F. Microbial and metabolic characterization of organic artisanal sauerkraut fermentation and study of gut health-promoting properties of sauerkraut brine. Front. Microbiol. 2022, 13, 929738. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Paramithiotis, S.; Shin, H.S. Kimchi and Other Widely Consumed Traditional Fermented Foods of Korea: A Review. Front. Microbiol. 2016, 7, 1493. [Google Scholar] [CrossRef]
- Perez-Diaz, I.M.; Hayes, J.; Medina, E.; Anekella, K.; Daughtry, K.; Dieck, S.; Levi, M.; Price, R.; Butz, N.; Lu, Z.; et al. Reassessment of the succession of lactic acid bacteria in commercial cucumber fermentations and physiological and genomic features associated with their dominance. Food Microbiol. 2017, 63, 217–227. [Google Scholar] [CrossRef]
- Stoll, D.A.; Muller, A.; Meinhardt, A.K.; Dotsch, A.; Greiner, R.; Kulling, S.E.; Huch, M. Influence of salt concentration and iodized table salt on the microbiota of fermented cucumbers. Food Microbiol. 2020, 92, 103552. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, S.; Bhattacharya, S.; Chatzinotas, A.; Chakraborty, W.; Bhattacharya, D.; Gachhui, R. Kombucha tea fermentation: Microbial and biochemical dynamics. Int. J. Food Microbiol. 2016, 220, 63–72. [Google Scholar] [CrossRef]


| Sample | Source | Ingredients |
|---|---|---|
| Kombucha | ||
| 20 | Humm Kombucha | Water, raw sugar, black tea, and living kombucha culture |
| 21 | Renee Voltaire | Water, raw sugar cane, green tea (chun-mee), and kombucha culture |
| 1 | Individual 2 | 2 black teas, 1 oolong tea, sugar cane, and SCOBY (mid–late December) |
| 2 | Individual 2 | 2 black teas, 1 oolong tea, sugar cane and SCOBY (late December–mid January) |
| 3 | Individual 2 | 2 black teas, 1 oolong tea, sugar cane and SCOBY (mid–end of January) |
| Water Kefir | ||
| 28 | Renee Voltaire | Water, agave, sugar, lemon juice, ginger, and living water kefir culture (L. lactis lactis, L. lactis cremoris, L. lactis diacetylactis, L. casei, L. acidophilus, and Kluyveromyces marxianus (yeast)) |
| 29 | Grönt Levande | Water, raw sugar, water kefir grains, ginger, lime, and figs |
| 30 | Kombucheriet | Water, living water kefir culture, raw sugar, and rosehip |
| 31 | Kombucheriet | Water, living water kefir culture, raw sugar, and rhubarb juice |
| 26 | Individual 1 | Water, muscovado sugar, and water kefir grains (3 days). Same batch of water kefir grains as Sample 27 |
| 27 | Individual 1 | Water, muscovado sugar, and water kefir grains (4 days). Same batch of water kefir grains as Sample 26 |
| Milk Kefir | ||
| 23 | Valio | Pasteurized milk, lactase enzyme, starter culture (bacteria and yeast), L. rhamnosus GG, and vitamin D (2,5% fat) |
| 24 | Arla | Pasteurized milk, starter culture, L. acidophilus LA-5, B. lactis BB-12, L. casei F-19 and vitamin D (3.0% fat) |
| 25 | Individual 1 | Milk (Garant eko 3% fat) and starter culture from yo*gut; made on the same date as Sample 8 |
| 8 | Individual 1 | Milk (Garant eko 3% fat) and milk kefir grains from yo*gut (see legend for microbial composition*); same date as Sample 25 |
| 9 | Individual 1 | Milk (Garant eko 3% fat) and milk kefir grains from yo*gut (see legend for microbial composition*); milk kefir grains from same batch as Sample 10 |
| 10 | Individual 1 | Milk (Garant eko 3% fat) and milk kefir grains from yo*gut shop (see legend for microbial composition*); milk kefir grains from same batch as Sample 9 |
| Yogurt (plain) | ||
| 32 | Milchbauern Schrozberger | Milk (3.5%), skimmed milk powder, living bacteria culture, L. acidophilus, and B. bifidum |
| 33 | Wapnö | Pasteurized milk, cream, and yogurt culture (8% fat, total) |
| 34 | Larsa Foods | Homogenized and pasteurized milk (0%) and yogurt culture |
| 35 | Individual 1 | Milk (Arla 3% fat) and bacterial culture blend from YOGUT ME |
| 36 | Individual 1 | Milk (Arla 3% fat) and bacterial culture from Yogourmet (L. delbrueckii bulgaricus, S. thermophilus, L. acidophilus) |
| 37 | Individual 1 | Milk (Arla 3% fat) and bacterial cultures from Cultures for Health |
| Plant-based yoghurt | ||
| 38 | My Love My Life | Water, almond, tapioca starch, stabilizer, locust bean gum, salt, and lactic acid culture |
| 39 | So Soja | Soya drink (organic) 99% and live cultures including Bifidobacterium and L. acidophilus |
| 40 | Harvest Moon | Coconut milk 99%, tapioca, yogurt culture, S. thermophilis, L. bulgaricus, L. acidophilus, and B. lactis |
| 41 | Individual 1 | Coconut milk, cashew nut, tapioca starch, and YOGUT ME culture blend |
| 42 | Individual 1 | Coconut milk and YOGUT ME culture blend (batch 1) |
| 48 | Individual 1 | Coconut milk and YOGUT ME culture blend (batch 2) |
| 43 | Individual 1 | Coconut milk and Belle and Bella yoghurt culture |
| Sauerkraut | ||
| 11 | Grönt Levande | White cabbage, onion, salt, garlic, juniper berry, and mustard seeds |
| 12 | Ölands Ljuvliga | White cabbage and salt |
| 13 | Tistelvind | White cabbage, cumin, juniper berry, and sea salt |
| 5 | Individual 1 | White cabbage and salt (2%) |
| 6 | Individual 1 | White cabbage, turmeric, garlic, and salt (2%) |
| 7 | Individual 1 | Red cabbage and red onion |
| 4 | Individual 2 | White cabbage, caraway, bay leaves, and salt (2.5%). |
| Kimchi | ||
| 14 | Tistelvind | Savoy cabbage, carrot, daikon radish, onion, garlic, ginger, chili pepper, and sea salt (“hot”, jar 1) |
| 15 | Tistelvind | Savoy cabbage, carrot, daikon radish, onion, garlic, ginger, chili pepper, and sea salt (“hot”, jar 2) |
| 17 | Tistelvind | Savoy cabbage, carrot, daikon radish, onion, garlic, ginger, chili pepper, and sea salt (“medium hot”, jar 3) |
| 18 | Tistelvind | Savoy cabbage, carrot, daikon radish, onion, garlic, ginger, chili pepper, and sea salt (“medium hot”, jar 4) |
| 16 | Grönt Levande | White cabbage, onion, carrot, garlic, salt, radish, ginger, chili, water kefir, and bacterial culture |
| 19 | Individual 2 | White cabbage, spring onion, ginger, garlic, kimchi chili powder flakes, and salt |
| Fermented vegetables | ||
| 44 | Tistelvind | Beetroot, white cabbage and sea salt |
| 45 | Tistelvind | Carrot and sea salt |
| 46 | Grönt Levande | Cucumber and sea salt |
| 47 | Tistelvind | Cucumber and sea salt |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palmnäs-Bédard, M.; de Santa Izabel, A.; Dicksved, J.; Landberg, R. Characterization of the Bacterial Composition of 47 Fermented Foods in Sweden. Foods 2023, 12, 3827. https://doi.org/10.3390/foods12203827
Palmnäs-Bédard M, de Santa Izabel A, Dicksved J, Landberg R. Characterization of the Bacterial Composition of 47 Fermented Foods in Sweden. Foods. 2023; 12(20):3827. https://doi.org/10.3390/foods12203827
Chicago/Turabian StylePalmnäs-Bédard, Marie, Aline de Santa Izabel, Johan Dicksved, and Rikard Landberg. 2023. "Characterization of the Bacterial Composition of 47 Fermented Foods in Sweden" Foods 12, no. 20: 3827. https://doi.org/10.3390/foods12203827